Our group uses an interdisciplinary combination of molecular/cellular and computational methods to understand how disease susceptibility is encoded in the non-coding portion of the genome. Our focus is on type 2 diabetes (T2D) and related traits. Looking forward, our belief is that high-throughput biological profiling and analysis approaches will be closely tied to disease diagnosis, prognosis, and treatment — and will therefore have a tremendous influence on medicine.
We generate and computationally analyze multiple high-throughput data sets on the genome (DNA-seq), epigenome (ATAC-seq), transcriptome (RNA-seq), and metabolome in disease-relevant cells and tissues. Our target tissues of interest are pancreatic islets, skeletal muscle, and adipose. Computational integration of these data sets with publically-available data sets from other cells and tissues allows us to discover tissue-specific and disease-relevant functional signatures.
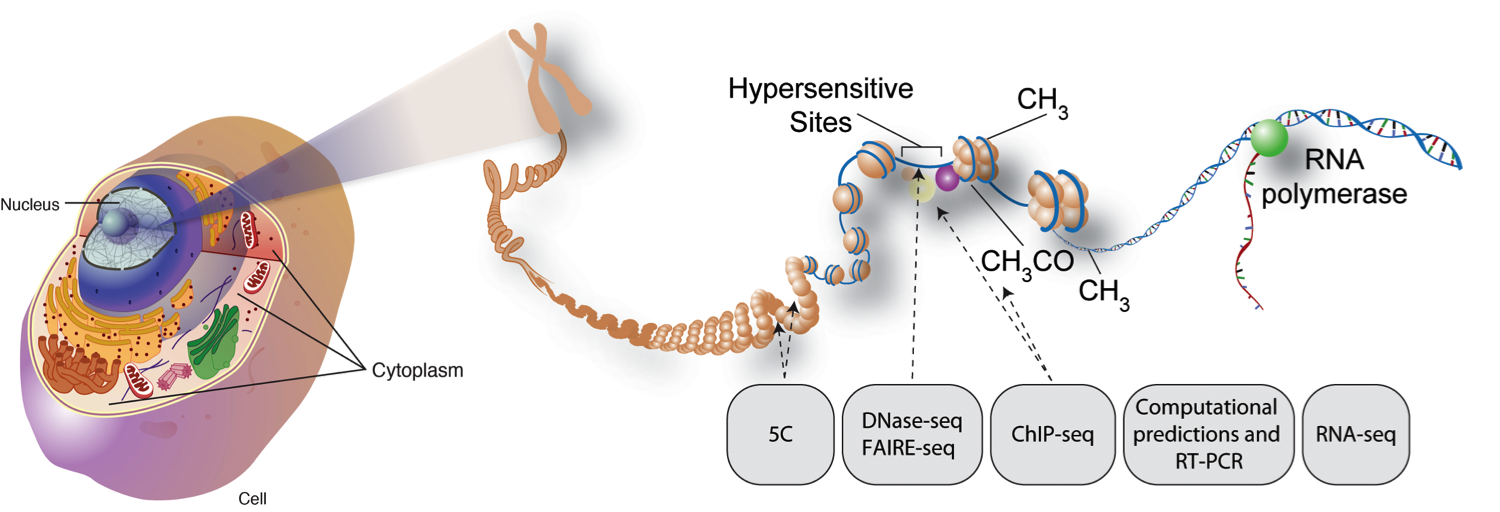
Eukaryotic chromatin architecture.
SourceOur high-throughput data sets are generated across multiple species - currently in human, mouse, and rat. We develop and use comparative approaches to integrate these data sets, which allows for understanding of the evolutionary processes and translation of our model species maps at orthologous human loci.

Genome alignment.
SourceIn addition to generating and analyzing cross-species data sets, we generate and analyze molecular profiles across a diversity of human subjects. Integrating genome and functional profiles allows us to observe the effect of genetic variation on non-coding functional regions. Together, these findings will illuminate the basic molecular mechanisms encoding T2D and related trait susceptibility and will provide new insights into the foundations of metabolic health and longevity.
Genome alignment.
Source