RFX6-mediated dysregulation defines human β cell dysfunction in early type 2 diabetes
Highlights
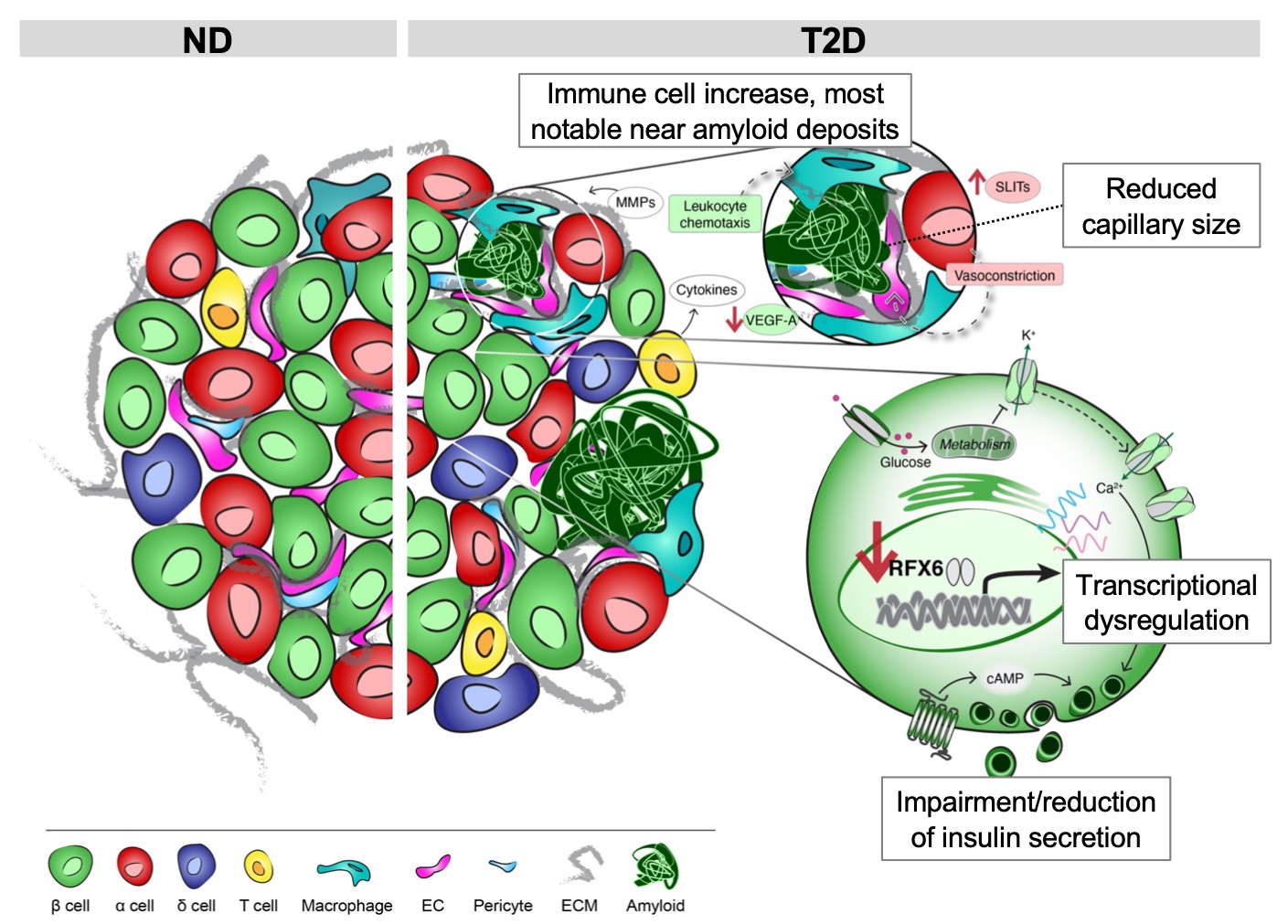
- Physiological, transcriptomic, and image profiling of short-duration T2D and ND pancreatic islets
- Changes in islet microenvironment changes and reduced β cell function define key features of early-stage T2D
- Co-expression network analyses identified gene modules related to donor and islet traits and revealed disrupted metabolism and cilia homeostasis in T2D
- β cell hub gene RFX6 is reduced in T2D and controls glucose-stimulated insulin secretion
- RFX6 knockdown alters the β cell chromatin and transcriptional landscape and downregulates secretory vesicle components
Abstract
A hallmark of type 2 diabetes (T2D) is dysfunction of insulin-producing β cells in islets of Langerhans. T2D genome-wide association studies (GWAS) have identified hundreds of signals but translating these into biological mechanisms is challenging. To identify early disease-driving events, we performed integrated studies of short-duration T2D and control donors including pancreatic tissue single cell spatial proteomics and islet physiology and transcriptomics. We show that T2D is associated with β cell dysfunction, but not β cell loss, and define cellular neighborhood organizational changes in intraislet capillaries and T cells. We found that a transcription factor RFX6 gene regulatory network is associated with insulin secretion defects and T2D GWAS variants, and further validated the RFX6 role in β cells through direct perturbation in human pseudoislets with physiological and single nuclei multiome readouts. Thus, β cell-intrinsic defects, including RFX6-mediated β cell chromatin, transcriptome, and insulin secretion dysregulation, define early-stage T2D signatures.
Important links
- Co-expression network modules: R Shiny app to explore celltype WGCNA modules
- Pseudoislet single-cell multiome: Explore single-cell data from RFX6 Kd/Ctrl multiome profiling experiment
- Pancreatlas: Browse immunofluorescence and CODEX images used for spatial analyses
Code and data availability
EGA submission is in progress. Source code will be made available on GitHub.
Lead authors
John T. Walker
Diane C. Saunders
Vivek Rai
Corresponding authors
Steve Parker
University of Michigan
Marcela Brissova
Vanderbilt University
Alvin C. Powers
Vanderbilt University
